MMACE Paper: Recurrent Neural Network for Predicting Solubility
Show code cell source
# import os
# os.environ["CUDA_VISIBLE_DEVICES"] = "0"
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib as mpl
import numpy as np
import tensorflow as tf
import selfies as sf
import exmol
from dataclasses import dataclass
from rdkit.Chem.Draw import rdDepictor
rdDepictor.SetPreferCoordGen(True)
sns.set_context("notebook")
sns.set_style(
"dark",
{
"xtick.bottom": True,
"ytick.left": True,
"xtick.color": "#666666",
"ytick.color": "#666666",
"axes.edgecolor": "#666666",
"axes.linewidth": 0.8,
"figure.dpi": 300,
},
)
color_cycle = ["#1BBC9B", "#F06060", "#5C4B51", "#F3B562", "#6e5687"]
mpl.rcParams["axes.prop_cycle"] = mpl.cycler(color=color_cycle)
soldata = pd.read_csv(
"https://github.com/whitead/dmol-book/raw/main/data/curated-solubility-dataset.csv"
)
features_start_at = list(soldata.columns).index("MolWt")
np.random.seed(0)
2025-05-08 18:30:39.774229: I external/local_xla/xla/tsl/cuda/cudart_stub.cc:32] Could not find cuda drivers on your machine, GPU will not be used.
2025-05-08 18:30:39.777619: I external/local_xla/xla/tsl/cuda/cudart_stub.cc:32] Could not find cuda drivers on your machine, GPU will not be used.
2025-05-08 18:30:39.786111: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:467] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
E0000 00:00:1746729039.800694 20005 cuda_dnn.cc:8579] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered
E0000 00:00:1746729039.804958 20005 cuda_blas.cc:1407] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
W0000 00:00:1746729039.816897 20005 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1746729039.816918 20005 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1746729039.816920 20005 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
W0000 00:00:1746729039.816921 20005 computation_placer.cc:177] computation placer already registered. Please check linkage and avoid linking the same target more than once.
2025-05-08 18:30:39.821243: I tensorflow/core/platform/cpu_feature_guard.cc:210] This TensorFlow binary is optimized to use available CPU instructions in performance-critical operations.
To enable the following instructions: AVX2 FMA, in other operations, rebuild TensorFlow with the appropriate compiler flags.
# REDUCED Data FOR CI
soldata = soldata.sample(frac=0.1, random_state=0).reset_index(drop=True)
soldata.head()
| ID | Name | InChI | InChIKey | SMILES | Solubility | SD | Ocurrences | Group | MolWt | ... | NumRotatableBonds | NumValenceElectrons | NumAromaticRings | NumSaturatedRings | NumAliphaticRings | RingCount | TPSA | LabuteASA | BalabanJ | BertzCT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | B-4206 | diuron | InChI=1S/C9H10Cl2N2O/c1-13(2)9(14)12-6-3-4-7(1... | XMTQQYYKAHVGBJ-UHFFFAOYSA-N | CN(C)C(=O)Nc1ccc(Cl)c(Cl)c1 | -3.744300 | 1.227164 | 5 | G4 | 233.098 | ... | 1.0 | 76.0 | 1.0 | 0.0 | 0.0 | 1.0 | 32.34 | 92.603980 | 2.781208 | 352.665233 |
| 1 | F-988 | 7-(3-amino-3-methylazetidin-1-yl)-8-chloro-1-c... | InChI=1S/C17H17ClFN3O3/c1-17(20)6-21(7-17)14-1... | DUNZFXZSFJLIKR-UHFFFAOYSA-N | CC1(N)CN(C2=C(Cl)C3=C(C=C2F)C(=O)C(C(=O)O)=CN3... | -5.330000 | 0.000000 | 1 | G1 | 365.792 | ... | 3.0 | 132.0 | 2.0 | 2.0 | 2.0 | 4.0 | 88.56 | 147.136366 | 2.001398 | 973.487509 |
| 2 | C-1996 | 4-acetoxybiphenyl; 4-biphenylyl acetate | InChI=1S/C14H12O2/c1-11(15)16-14-9-7-13(8-10-1... | MISFQCBPASYYGV-UHFFFAOYSA-N | CC(=O)OC1=CC=C(C=C1)C2=CC=CC=C2 | -4.400000 | 0.000000 | 1 | G1 | 212.248 | ... | 2.0 | 80.0 | 2.0 | 0.0 | 0.0 | 2.0 | 26.30 | 94.493449 | 2.228677 | 471.848345 |
| 3 | A-3055 | methane dimolybdenum | InChI=1S/CH4.2Mo/h1H4;; | JAGQSESDQXCFCH-UHFFFAOYSA-N | C.[Mo].[Mo] | -3.420275 | 0.409223 | 2 | G3 | 207.923 | ... | 0.0 | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.00 | 49.515427 | -0.000000 | 2.754888 |
| 4 | A-2575 | ethyl 4-[[(methylphenylamino)methylene]amino]b... | InChI=1S/C17H18N2O2/c1-3-21-17(20)14-9-11-15(1... | GNGYPJUKIKDJQT-UHFFFAOYSA-N | CCOC(=O)c1ccc(cc1)N=CN(C)c2ccccc2 | -5.450777 | 0.000000 | 1 | G1 | 282.343 | ... | 5.0 | 108.0 | 2.0 | 0.0 | 0.0 | 2.0 | 41.90 | 124.243431 | 2.028889 | 606.447052 |
5 rows × 26 columns
selfies_list = []
for s in soldata.SMILES:
try:
selfies_list.append(sf.encoder(exmol.sanitize_smiles(s)[1]))
except sf.EncoderError:
selfies_list.append(None)
len(selfies_list)
998
basic = set(exmol.get_basic_alphabet())
data_vocab = set(
sf.get_alphabet_from_selfies([s for s in selfies_list if s is not None])
)
vocab = ["[nop]"]
vocab.extend(list(data_vocab.union(basic)))
vocab_stoi = {o: i for o, i in zip(vocab, range(len(vocab)))}
def selfies2ints(s):
result = []
for token in sf.split_selfies(s):
if token == ".":
continue # ?
if token in vocab_stoi:
result.append(vocab_stoi[token])
else:
result.append(np.nan)
# print('Warning')
return result
def ints2selfies(v):
return "".join([vocab[i] for i in v])
# test them out
s = selfies_list[0]
print("selfies:", s)
v = selfies2ints(s)
print("selfies2ints:", v)
so = ints2selfies(v)
print("ints2selfes:", so)
assert so == s.replace(
".", ""
) # make sure '.' is removed from Selfies string during assertion
selfies: [C][N][Branch1][C][C][C][=Branch1][C][=O][N][C][=C][C][=C][Branch1][C][Cl][C][Branch1][C][Cl][=C][Ring1][Branch2]
selfies2ints: [6, 11, 69, 6, 6, 6, 5, 6, 19, 11, 6, 30, 6, 30, 69, 6, 36, 6, 69, 6, 36, 30, 16, 7]
ints2selfes: [C][N][Branch1][C][C][C][=Branch1][C][=O][N][C][=C][C][=C][Branch1][C][Cl][C][Branch1][C][Cl][=C][Ring1][Branch2]
@dataclass
class Config:
vocab_size: int
example_number: int
batch_size: int
buffer_size: int
embedding_dim: int
rnn_units: int
hidden_dim: int
config = Config(
vocab_size=len(vocab),
example_number=len(selfies_list),
batch_size=16,
buffer_size=10000,
embedding_dim=256,
hidden_dim=128,
rnn_units=128,
)
# now get sequences
encoded = [selfies2ints(s) for s in selfies_list if s is not None]
padded_seqs = tf.keras.preprocessing.sequence.pad_sequences(encoded, padding="post")
# Now build dataset
data = tf.data.Dataset.from_tensor_slices(
(padded_seqs, soldata.Solubility.iloc[[bool(s) for s in selfies_list]].values)
)
# now split into val, test, train and batch
N = len(data)
split = int(0.1 * N)
test_data = data.take(split).batch(config.batch_size)
nontest = data.skip(split)
val_data, train_data = nontest.take(split).batch(config.batch_size), nontest.skip(
split
).shuffle(config.buffer_size).batch(config.batch_size).prefetch(
tf.data.experimental.AUTOTUNE
)
2025-05-08 18:30:43.859276: E external/local_xla/xla/stream_executor/cuda/cuda_platform.cc:51] failed call to cuInit: INTERNAL: CUDA error: Failed call to cuInit: UNKNOWN ERROR (303)
model = tf.keras.Sequential()
# make embedding and indicate that 0 should be treated as padding mask
model.add(
tf.keras.layers.Embedding(
input_dim=config.vocab_size, output_dim=config.embedding_dim, mask_zero=True
)
)
# RNN layer
model.add(tf.keras.layers.GRU(config.rnn_units))
# a dense hidden layer
model.add(tf.keras.layers.Dense(config.hidden_dim, activation="relu"))
# regression, so no activation
model.add(tf.keras.layers.Dense(1))
model.summary()
Model: "sequential"
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓ ┃ Layer (type) ┃ Output Shape ┃ Param # ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩ │ embedding (Embedding) │ ? │ 0 (unbuilt) │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ gru (GRU) │ ? │ 0 (unbuilt) │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ dense (Dense) │ ? │ 0 (unbuilt) │ ├─────────────────────────────────┼────────────────────────┼───────────────┤ │ dense_1 (Dense) │ ? │ 0 (unbuilt) │ └─────────────────────────────────┴────────────────────────┴───────────────┘
Total params: 0 (0.00 B)
Trainable params: 0 (0.00 B)
Non-trainable params: 0 (0.00 B)
model.compile(tf.optimizers.Adam(1e-4), loss="mean_squared_error")
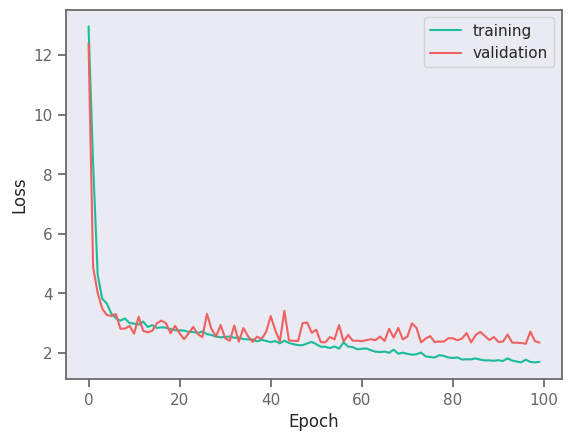
result = model.fit(train_data, validation_data=val_data, epochs=100, verbose=0)
plt.plot(result.history["loss"], label="training")
plt.plot(result.history["val_loss"], label="validation")
plt.legend()
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.show()
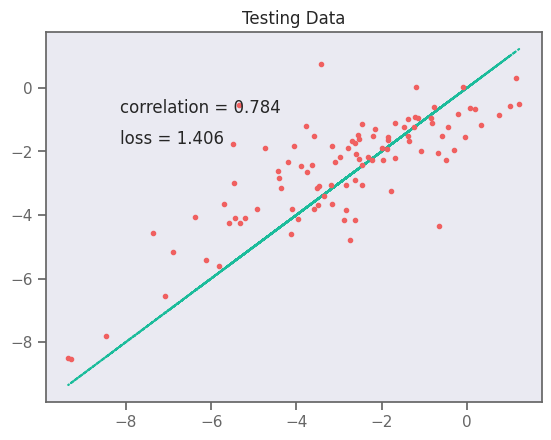
yhat = []
test_y = []
for x, y in test_data:
yhat.extend(model(x).numpy().flatten())
test_y.extend(y.numpy().flatten())
yhat = np.array(yhat)
test_y = np.array(test_y)
# plot test data
plt.plot(test_y, test_y, ":")
plt.plot(test_y, yhat, ".")
plt.text(min(y) - 7, max(y) - 2, f"correlation = {np.corrcoef(test_y, yhat)[0,1]:.3f}")
plt.text(min(y) - 7, max(y) - 3, f"loss = {np.sqrt(np.mean((test_y - yhat)**2)):.3f}")
plt.title("Testing Data")
plt.savefig("rnn-fit.png", dpi=300)
plt.show()
2025-05-08 18:37:39.612676: I tensorflow/core/framework/local_rendezvous.cc:407] Local rendezvous is aborting with status: OUT_OF_RANGE: End of sequence
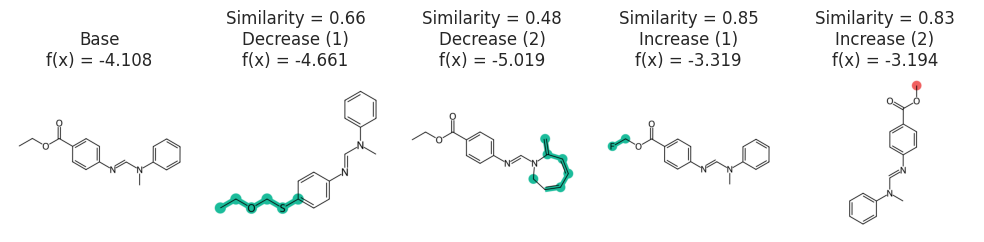
CF explanation:
In the following example let’s say we would like our molecules to return a solubility value of -3.5. Here we use MMACE algorithm to createcounter factual explanations. In other words, we would like to see what are the minimal mutations that could to be done to our input structure to get our desired solubility.
def predictor_function(smile_list, selfies):
encoded = [selfies2ints(s) for s in selfies]
# check for nans
valid = [1.0 if sum(e) > 0 else np.nan for e in encoded]
encoded = [np.nan_to_num(e, nan=0) for e in encoded]
padded_seqs = tf.keras.preprocessing.sequence.pad_sequences(encoded, padding="post")
labels = np.reshape(model.predict(padded_seqs, verbose=0), (-1))
return labels * valid
predictor_function([], ["[C][C][O]", "[C][C][Nop][O]"])
array([0.26268968, nan])
stoned_kwargs = {
"num_samples": 2500,
"alphabet": exmol.get_basic_alphabet(),
"max_mutations": 2,
}
space = exmol.sample_space(
soldata.SMILES[4], predictor_function, stoned_kwargs=stoned_kwargs, quiet=True
)
exps = exmol.rcf_explain(space, 0.5, nmols=4)
fkw = {"figsize": (10, 3)}
exmol.plot_cf(exps, figure_kwargs=fkw, mol_size=(450, 400), nrows=1)
plt.savefig("rnn-simple.png", bbox_inches="tight", dpi=180)
svg = exmol.insert_svg(exps, mol_fontsize=16)
with open("rnn-simple.svg", "w") as f:
f.write(svg)
fkw = {"figsize": (10, 4)}
font = {"family": "normal", "weight": "normal", "size": 22}
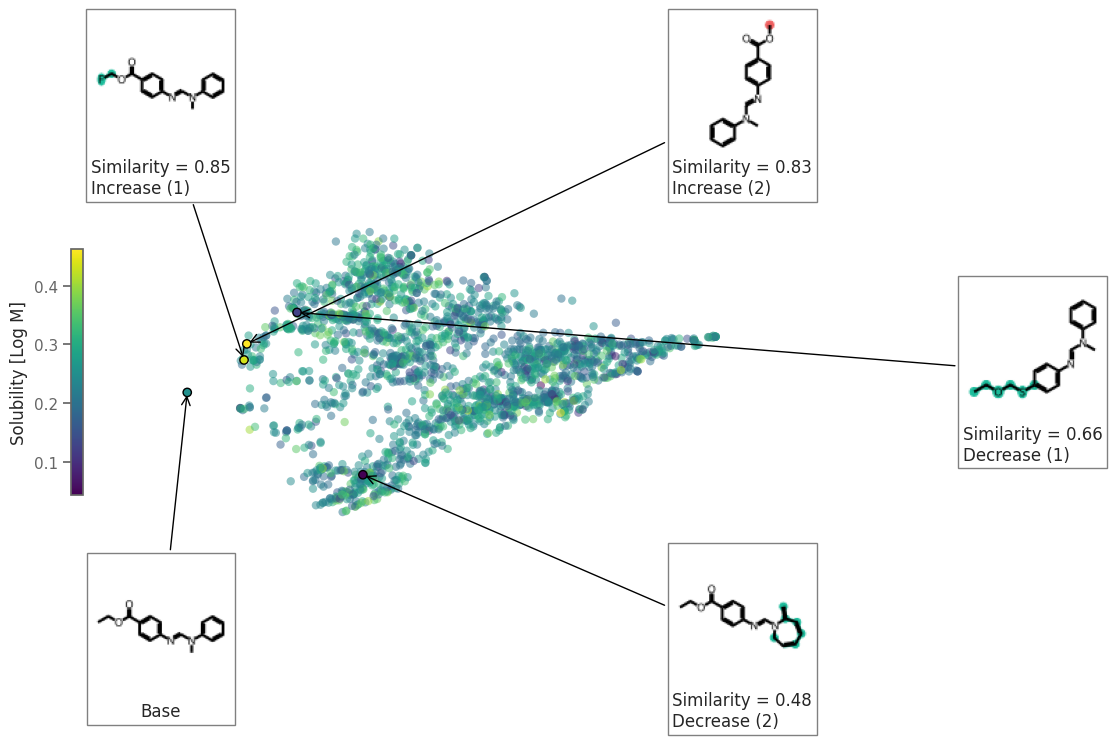
exmol.plot_space(space, exps, figure_kwargs=fkw, mol_size=(100, 100), offset=1)
ax = plt.gca()
plt.colorbar(
ax.get_children()[1],
ax=[ax],
label="Solubility [Log M]",
location="left",
shrink=0.8,
)
plt.savefig("rnn-space.png", bbox_inches="tight", dpi=180)
svg = exmol.insert_svg(exps, mol_fontsize=16)
with open("svg_figs/rnn-space.svg", "w") as f:
f.write(svg)
space = exmol.sample_space(
soldata.SMILES[4], predictor_function, preset="wide", quiet=True
)
exps = exmol.rcf_explain(space, 0.5)
fkw = {"figsize": (8, 6)}
font = {"family": "normal", "weight": "normal", "size": 22}
exmol.plot_space(space, exps, figure_kwargs=fkw, mol_size=(200, 200), offset=1)
ax = plt.gca()
plt.colorbar(ax.get_children()[1], ax=[ax], location="left", label="Solubility [Log M]")
plt.savefig("rnn-wide.png", bbox_inches="tight", dpi=180)
svg = exmol.insert_svg(exps, mol_fontsize=16)
with open("rnn-space-wide.svg", "w") as f:
f.write(svg)
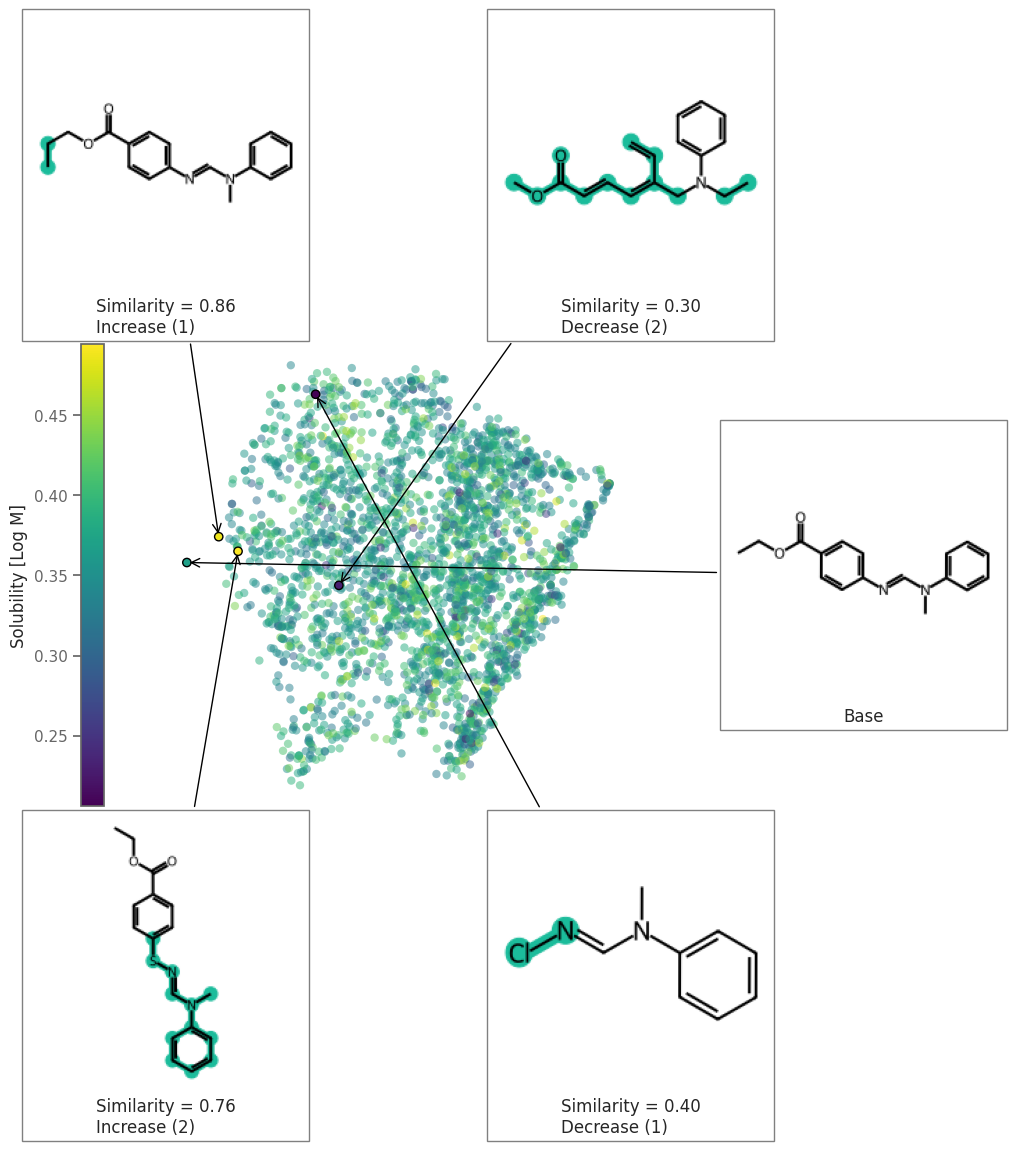
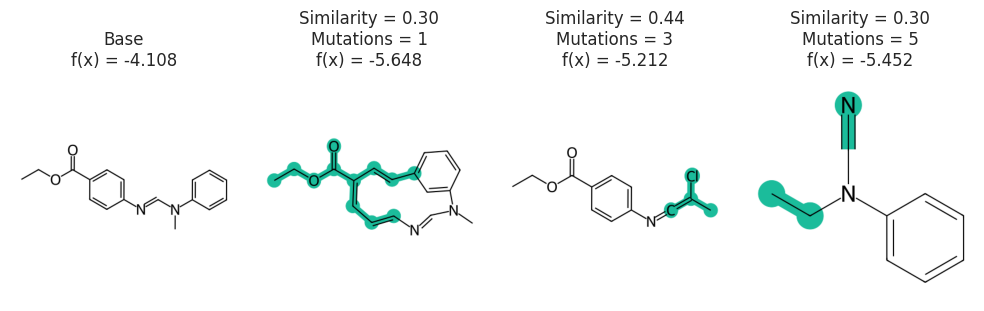
Figure showing effect of mutation number and Alphabet
exps = []
spaces = []
for i in [1, 3, 5]:
stoned_kwargs = {
"num_samples": 2500,
"alphabet": exmol.get_basic_alphabet(),
"min_mutations": i,
"max_mutations": i,
}
space = exmol.sample_space(
soldata.SMILES[4], predictor_function, stoned_kwargs=stoned_kwargs, quiet=True
)
spaces.append(space)
e = exmol.rcf_explain(space, nmols=2)
if len(exps) == 0:
exps.append(e[0])
for ei in e:
if not ei.is_origin and "Decrease" in ei.label:
ei.label = f"Mutations = {i}"
exps.append(ei)
break
fkw = {"figsize": (10, 4)}
exmol.plot_cf(exps, figure_kwargs=fkw, mol_fontsize=26, mol_size=(400, 400), nrows=1)
plt.savefig("rnn-mutations.png", bbox_inches="tight", dpi=180)
svg = exmol.insert_svg(exps, mol_fontsize=16)
with open("rnn-mutations.svg", "w") as f:
f.write(svg)
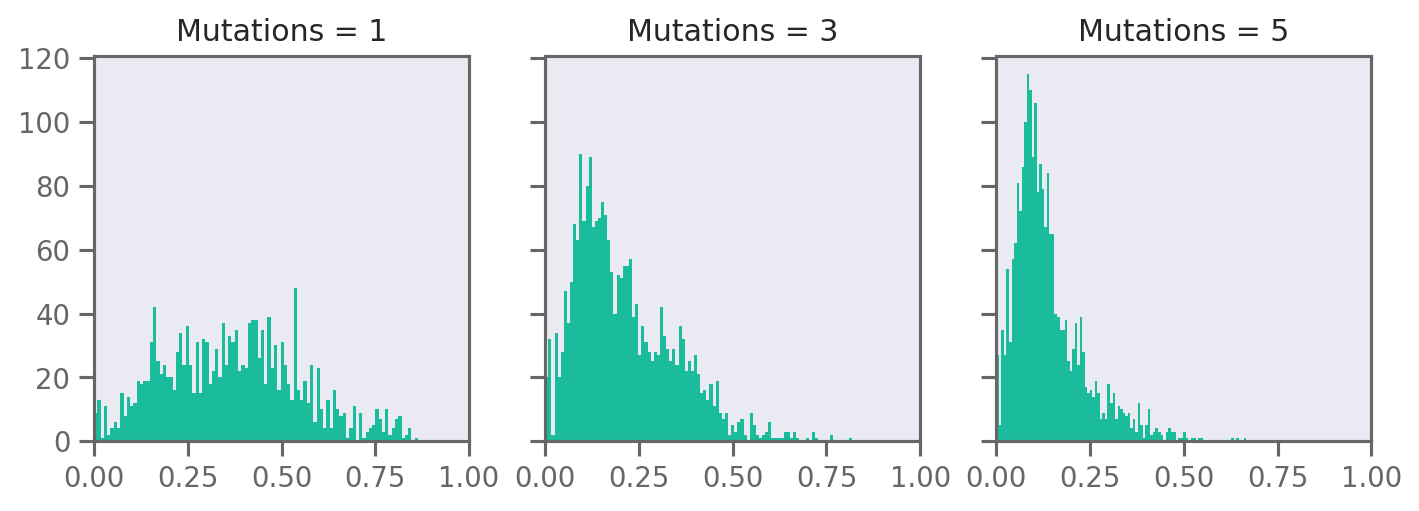
fig, axs = plt.subplots(1, 3, figsize=(8, 3), dpi=180, squeeze=True, sharey=True)
for i, n in enumerate([1, 3, 5]):
axs[i].hist([e.similarity for e in spaces[i][1:]], bins=99, edgecolor="none")
axs[i].set_title(f"Mutations = {n}")
axs[i].set_xlim(0, 1)
plt.tight_layout()
plt.savefig("rnn-mutation-hist.png", bbox_inches="tight", dpi=180)
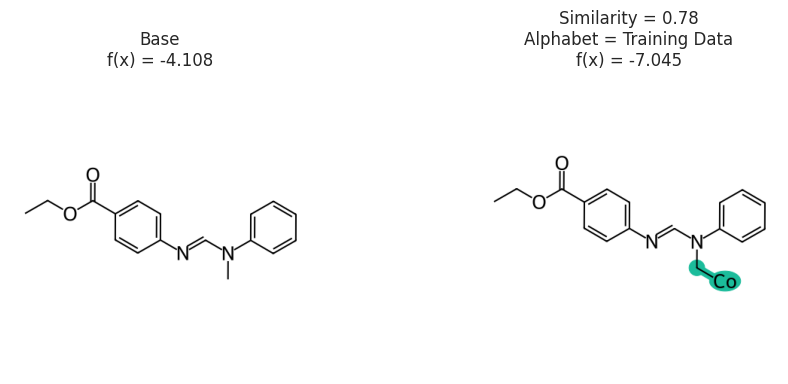
basic = exmol.get_basic_alphabet()
train = sf.get_alphabet_from_selfies([s for s in selfies_list if s is not None])
wide = sf.get_semantic_robust_alphabet()
alphs = {"Basic": basic, "Training Data": train, "SELFIES": wide}
exps = []
for l, a in alphs.items():
stoned_kwargs = {"num_samples": 2500 // 2, "alphabet": a, "max_mutations": 2}
space = exmol.sample_space(
soldata.SMILES[4], predictor_function, stoned_kwargs=stoned_kwargs, quiet=True
)
e = exmol.rcf_explain(space, nmols=2)
if len(exps) == 0:
exps.append(e[0])
for ei in e:
if not ei.is_origin and "Decrease" in ei.label:
ei.label = f"Alphabet = {l}"
exps.append(ei)
break
fkw = {"figsize": (10, 4)}
exmol.plot_cf(exps, figure_kwargs=fkw, mol_fontsize=26, mol_size=(400, 400), nrows=1)
plt.savefig("rnn-alphabets.png", bbox_inches="tight", dpi=180)
svg = exmol.insert_svg(exps, mol_fontsize=16)